Abstract
OtherAuthors: Muhammad Absar4, Tanveer Akhtar4, Masood Shammas8, Salman Basit9, Aamer Aleem10, Amer Mahmood10
8Dana Farbar Cancer Institute, University of Harvard , Boston USA, 9Centre for Genetics and Inherited Diseases, Taibah University, Madinah 10College of Medicine, King Saud University, Riyadh, Saudi Arabia.
Introduction:
Chronic Myeloid Leukemia (CML) results due to t(22;9) that leads to formation of Philadelphia chromosome and subsequently BCR-ABL fusion transcript, the later leading to leukemogenesis(1). This led to identification of tyrosine kinase inhibitors (TKIs) as novel drugs that revolutionised CML treatment in 21st century by improving the survival of CML equal to general population at least in developed countries (2). Nevertheless TKIs lose their effectiveness if CML patients progress to blast crisis (BC-CML). Despite extensive research in CML field, mechanism of its progression is still poorly understood and universal molecular biomarkers of CML progression are unknown (2).
There are different mechanisms associated with initiation and progression of various cancers. DNA repair defects mediated through biologically very important Fanconi anemia (FA) pathway genes are associated with many cancers (3). Nevertheless, FA pathway genes have not been reported in myeloid malignancies. In this study, in searching for mutated genes associated with many mechanism, we hereby report mutations in a very important FA-pathway gene associated exclusively with CML progression for the first time in literature.
Materials and Methods:
Patient selection: Whole-exome sequencing (WES) was employed to find out mutations in novel TF-genes associated with accelerated phase (AP-CML) and blast crisis phase CML (BC-CML) patients (Experimental groups 1 & 2, respectively). Chronic phase treatment-naïve CML patients (CP-CML) were used as control 1, CP-CML CML long-term TKI responders (at least 3 continuous years of MMR) as control group 2 and TK-resistant CML patients as control group 3, along with healthy controls.
Sample collection: DNA extraction and Clinical follow-up: 10 ml peripheral blood was collected from all study subjects. DNA was extracted and patient follow-up was carried out during course of this study. All treatment criteria per ENL guidelines were adopted.
Whole Exome Sequencing (WES): WES was carried out using Illumina NGS instrument (HiSeq). bcl files were converted to fastq files by using bcl2fastqtool (4). Raw reads were aligned to genome using BWA tools while whole exome variants were annotated using Illumina Variant Studio (4). R package was employed to align gene mutants to disease phenotypes (5). DNA repair genes mutated in all AP/BC-CML patients but not mutated in any of control groups were selected. Variants were confirmed using Sanger sequencing.
Results:
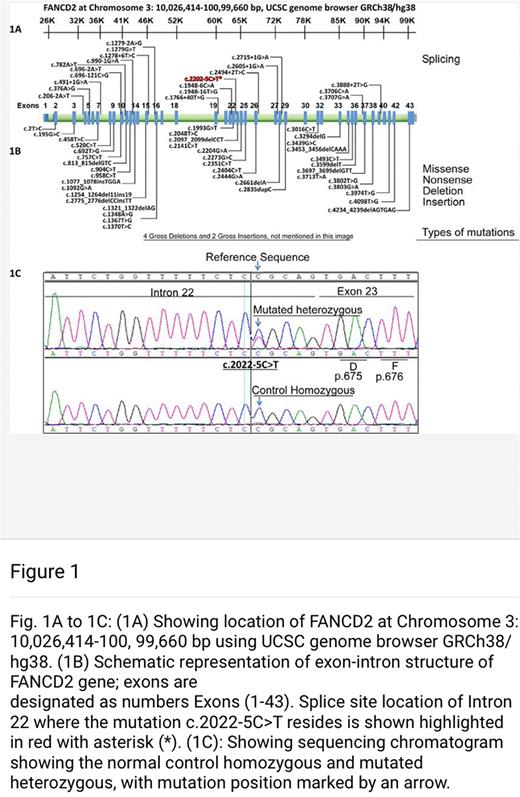
A novel splice-site substitution (C2022T) in FANCD2 gene (that is a very important member of FA-pathway genes) was detected in all AP/BC-CML but in none of the controls. It shows that mutated FANCD2 can serve as a novel biomarker exclusively for advanced phases of CML.
Discussion:
It is the first report that a member of FA-pathway genes (FANCD2) has been reported mutated in progression of myeloid malignancies (6). It shows that DNA repair defects and genomic instability mediated through FA-pathway could be one of the important mechanisms in CML progression. Mutations in FANCD2 and such other genes could be reason why CML blast crisis patients have poor outcome of stem cell transplantation. Moreover, anti-cancer drugs targeting DNA repair defects (PARP inhibitors) like Olaparib in combination with TKIs could be effective in patients with mutated FANCD-2 and other such genes.
Conflict of interest: The authors declare that they have no conflict of interests.
References:
1: Schwarz A, Roeder I, Seifert M. Cancers (Basel). 2022 Jan 5;14(1):256.
2: Annunziata M, Bonifacio M, Breccia M, Castagnetti F, Gozzini A, Iurlo A, et al. Front Oncol. 2020 ;10:883.
3: Nalepa G, Clapp DW. Nat Rev Cancer. 2018 Mar;18(3):168-185.
4: Hashmi JA, Albarry MA, Almatrafi AM, Albalawi AM, Mahmood A, Basit S. Congenit Anom (Kyoto). 2017 Apr 16. doi: 10.1111/cga.12225. [Epub ahead of print].
5: R Core Team (2012). R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R- project.org/
6: Chao MM, et al. Klin Padiatr. 2017 Nov;229(6):329-334.
7: Skelding KA, Lincz LF. Cancers (Basel). 2021 Oct 23;13(21):5328.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.